HiC 原理
A 3D Map of the Human Genome at Kilobase Resolution Reveals Principles of Chromatin Looping
- Hi-C protocol

- Chromatin architecture:
- Megadomains: 5–20 Mb intervals
- Topologically associated domains (TADs): ∼1 Mb
- Contact domains are often preserved across cell types: 40 kb to 3 Mb (median size 185 kb)
- Loops are short (<2 Mb) and strongly conserved across cell types and between human and mouse
- Eigenvector: Compartment A is highly enriched for open chromatin; compartment B is enriched for closed chromatin
- Genes whose promoters are associated with a loop are much more highly expressed than genes whose promoters are not associated with a loop (6-fold)
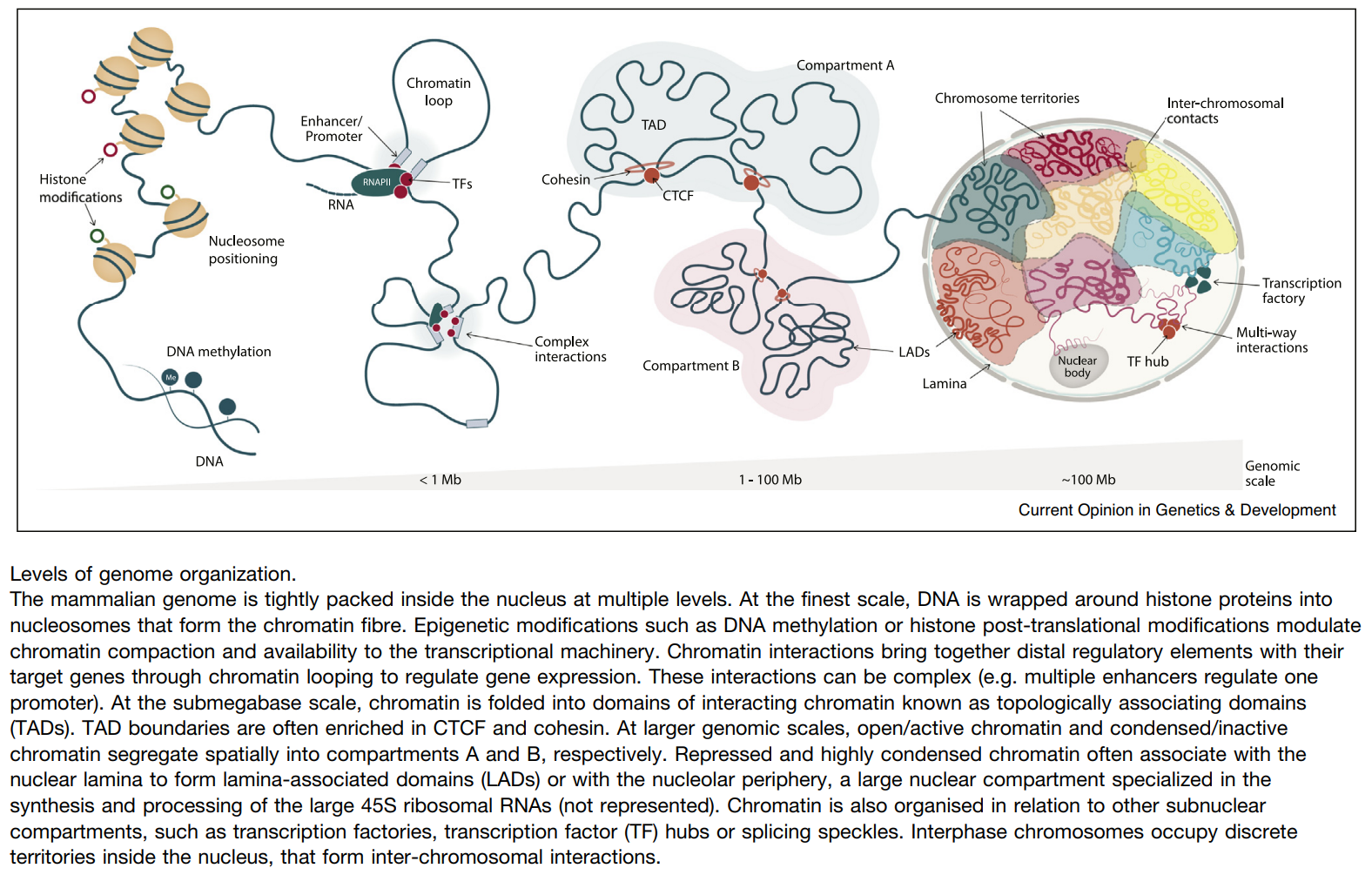 ref: Current Opinion in Genetics & Development, Izabela Harabula. 2021
ref: Current Opinion in Genetics & Development, Izabela Harabula. 2021
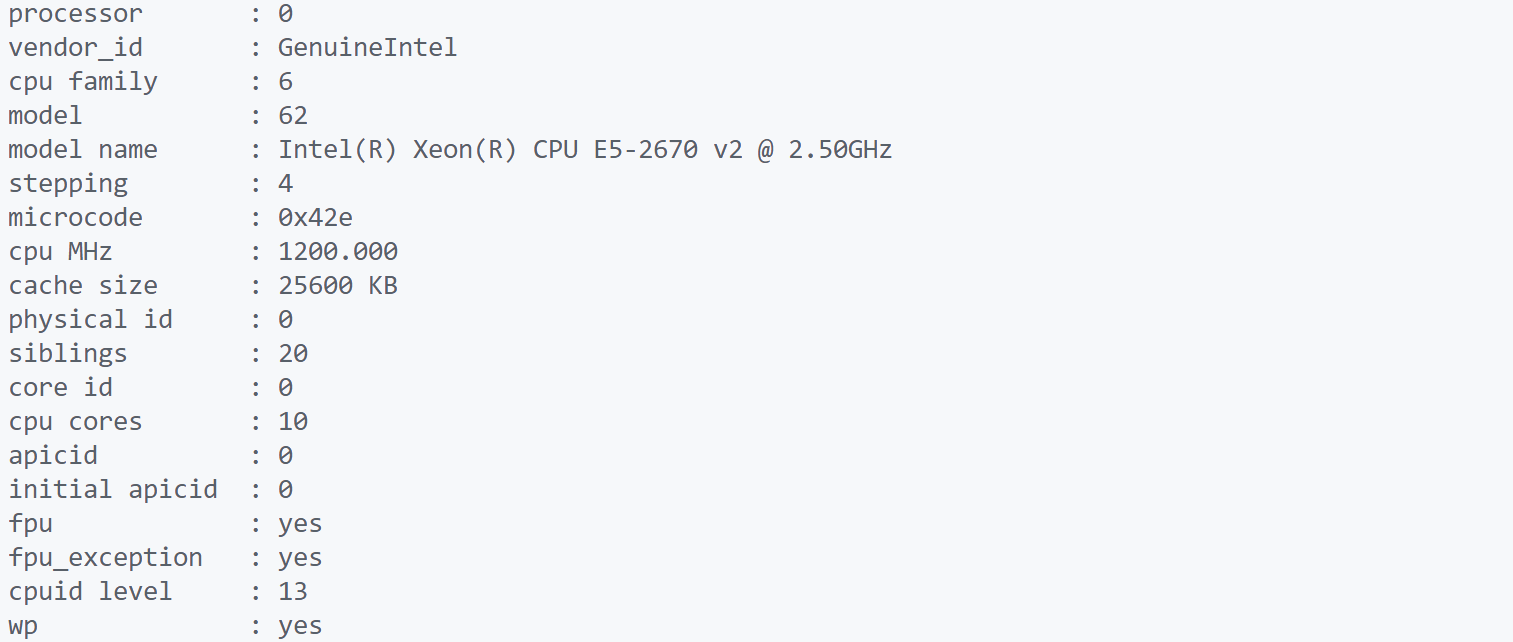
电脑配置信息
uname -a 查看内核/操作系统
#Linux chunyu-PowerEdge-R720 5.13.0-43-generic #48~20.04.1-Ubuntu SMP Thu May 12 12:59:19 UTC 2022 x86_64 x86_64 x86_64 GNU/Linux
df -h #硬盘信息
#文件系统 容量 已用 可用 已用% 挂载点
#rpool/USERDATA/lxh_gscj6g 46T 298G 46T 1% /home/lxh
#rpool/ROOT/ubuntu_ktv56z/usr/local 46T 3.7G 46T 1% /usr/local
free -h #内存信息
# 总计 已用 空闲 共享 缓冲/缓存 可用
#内存: 503Gi 260Gi 241Gi 10Mi 1.3Gi 240Gi
#交换: 2.0Gi 0B 2.0Gi
cat /proc/cpuinfo #CPU信息
cat /proc/cpuinfo| grep "physical id"| sort| uniq| wc -l #CPU数
cat /proc/cpuinfo | grep "cpu cores" | uniq #每个CPU物理核心数
cat /proc/cpuinfo| grep "processor"| wc -l #逻辑CPU数
nvidia-smi #查看GPU信息和使用情况
cut -d: -f1 /etc/passwd #查看系统所有用户
pstree -aps pid #查看进程关系
CPU信息展示:
GPU信息展示:
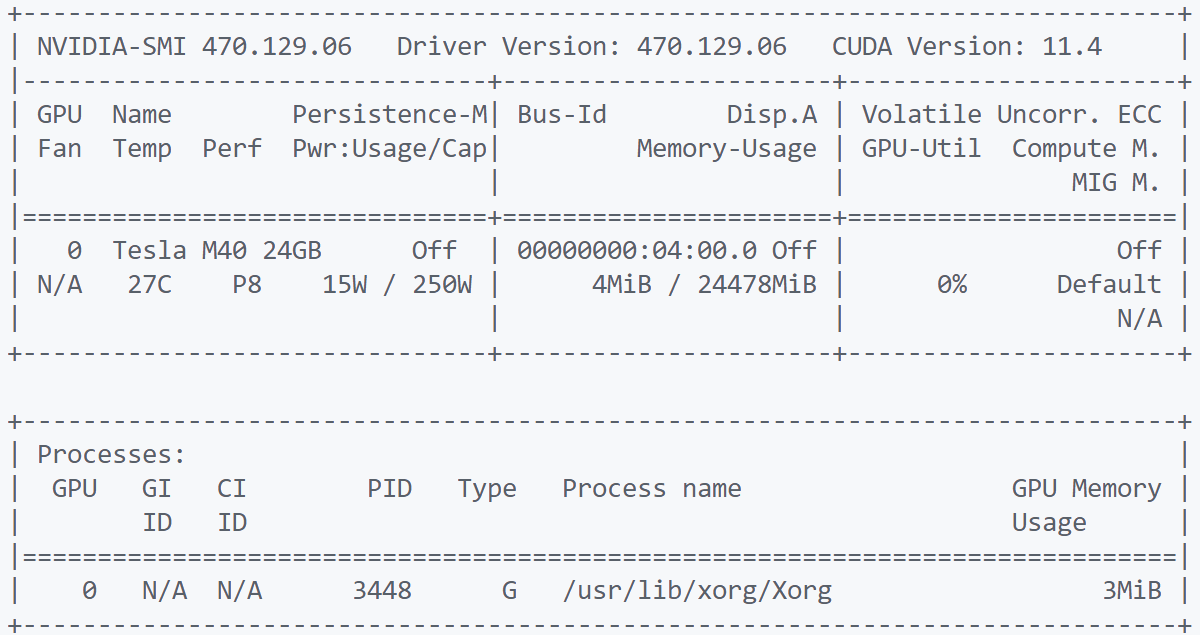
表头释义:
Fan:显示风扇转速,数值在0到100%之间,是计算机的期望转速,如果计算机不是通过风扇冷却或者风扇坏了,显示出来就是N/A;
Temp:显卡内部的温度,单位是摄氏度;
Perf:表征性能状态,从P0到P12,P0表示最大性能,P12表示状态最小性能;
Pwr:能耗表示;
Bus-Id:涉及GPU总线的相关信息;
Disp.A:是Display Active的意思,表示GPU的显示是否初始化;
Memory Usage:显存的使用率;
Volatile GPU-Util:浮动的GPU利用率;
Compute M:计算模式;
安装docker
参考教程:Install Docker Engine on Ubuntu
#Uninstall old versions
sudo apt-get remove docker docker-engine docker.io containerd runc
sudo rm -rf /var/lib/docker
sudo rm -rf /var/lib/containerd
##Install using the repository
#Set up the repository
sudo apt-get update
sudo apt-get install \
ca-certificates \
curl \
gnupg \
lsb-release
sudo mkdir -m 0755 -p /etc/apt/keyrings
curl -fsSL https://download.docker.com/linux/ubuntu/gpg | sudo gpg --dearmor -o /etc/apt/keyrings/docker.gpg
echo \
"deb [arch=$(dpkg --print-architecture) signed-by=/etc/apt/keyrings/docker.gpg] https://download.docker.com/linux/ubuntu \
$(lsb_release -cs) stable" | sudo tee /etc/apt/sources.list.d/docker.list > /dev/null
#Install Docker Engine
sudo apt-get update
sudo apt-get install docker-ce docker-ce-cli containerd.io docker-buildx-plugin docker-compose-plugin
sudo docker run hello-world
#添加用户进docker组,支持非root使用
sudo gpasswd -a username docker
newgrp docker
docker run hello-world
#GPU 支持(https://docs.nvidia.com/datacenter/cloud-native/container-toolkit/install-guide.html#installation-guide)
distribution=$(. /etc/os-release;echo $ID$VERSION_ID) \
&& curl -fsSL https://nvidia.github.io/libnvidia-container/gpgkey | sudo gpg --dearmor -o /usr/share/keyrings/nvidia-container-toolkit-keyring.gpg \
&& curl -s -L https://nvidia.github.io/libnvidia-container/$distribution/libnvidia-container.list | \
sed 's#deb https://#deb [signed-by=/usr/share/keyrings/nvidia-container-toolkit-keyring.gpg] https://#g' | \
sudo tee /etc/apt/sources.list.d/nvidia-container-toolkit.list
sudo apt-get update
sudo apt-get install -y nvidia-container-toolkit
sudo nvidia-ctk runtime configure --runtime=docker
sudo systemctl restart docker
sudo docker run --rm --runtime=nvidia --gpus all nvidia/cuda:11.6.2-base-ubuntu20.04 nvidia-smi
# apt-get install nvidia-container-runtime
CUDA and NVIDIA GPU介绍
创建hic_pipeline的docker环境
# hic-pipeline/
cd docker/hic-pipeline
docker build -t encodedcc/hic-pipeline:1.15.1 .
cd docker/delta
docker build -t encodedcc/hic-pipeline:1.15.1_delta .
cd docker/hiccups
docker build -t encodedcc/hic-pipeline:1.15.1_hiccups .
#或者从docker hub下载镜像
docker pull encodedcc/hic-pipeline:1.15.1
docker pull encodedcc/hic-pipeline:1.15.1_delta
docker pull encodedcc/hic-pipeline:1.15.1_hiccups
#列出安装的镜像
docker images
#REPOSITORY TAG IMAGE ID CREATED SIZE
#encodedcc/hic-pipeline 1.15.1_delta 41fc8e6dd680 2 hours ago 5.81GB
#encodedcc/hic-pipeline 1.15.1 95c8bf750530 3 months ago 2.21GB
#encodedcc/hic-pipeline 1.15.1_hiccups 363f736bd618 7 months ago 2.52GB
#hello-world latest feb5d9fea6a5 17 months ago 13.3kB
#启动容器
docker run -it encodedcc/hic-pipeline:1.15.1_hiccups /bin/bash
# -i 交互式使用
# -t 显示终端
# -v 主机路径:容器路径,实现主机和docker文件的共享,如 -v /home/lxh/Lab/ZheLiu/Hi-C/hic_run/hiccups_run/:/shared_folder
# --gpus all 将GPU添加到容器
#退出容器
exit
Juicer & Juicerbox
Juicer is a one-click pipeline for processing terabase scale Hi-C datasets.

基于Juicer的 ENCODE HiC-pipline 分析流程搭建
The ENCODE pipeline for processing Hi-C data based on Juicer.
简单使用
git clone https://github.com/ENCODE-DCC/hic-pipeline.git
cd hic-pipeline
pip install caper #依赖caper实现,首次运行时安装
caper run hic.wdl -i tests/functional/json/test_hic.json --docker
使用指南
-
JSON配置文件
参考Workflows说明 -
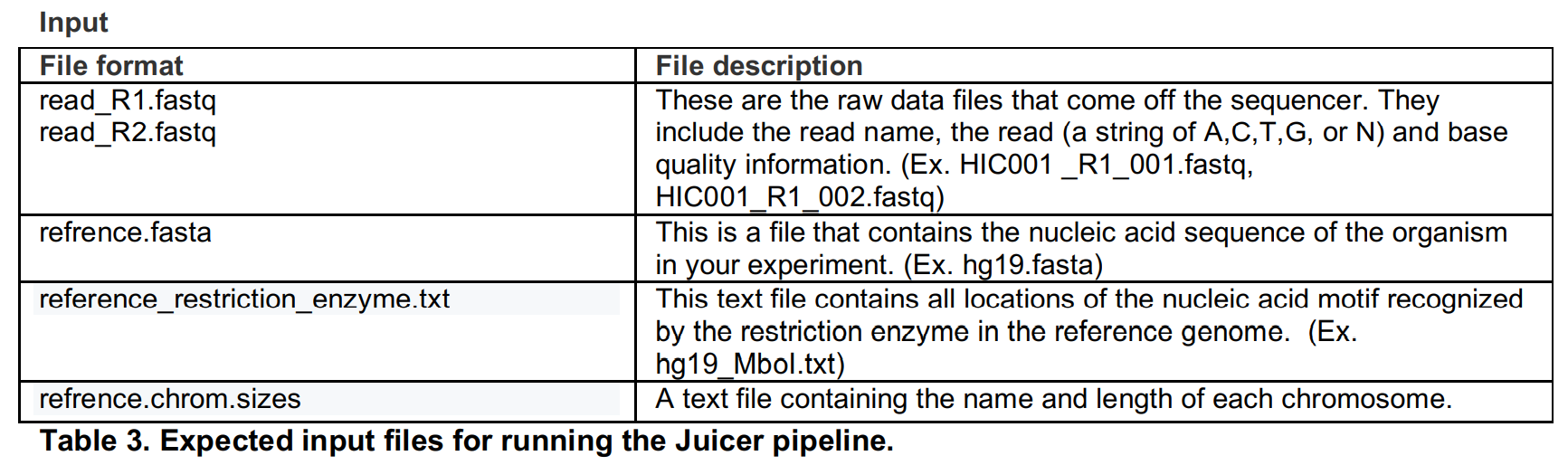
输入文件
需要准备文件包括测序数据,参考基因组序列文件,参考基因组bwa索引文件,参考基因组染色体大小文件,参考基因组酶切片段坐标信息文件。
hic.wdl的json配置文件内容如下:
{
"hic.assembly_name": "ce10",
"hic.reference_index": "tests/data/ce10_selected.tar.gz",
"hic.chrsz": "tests/data/ce10_selected.chrom.sizes.tsv",
"hic.reference_fasta": "",
"hic.restriction_enzymes": ["MboI"], #MboI, HindIII, DpnII, and none
"hic.restriction_sites": "tests/data/ce10_selected_MboI.txt.gz",
"hic.ligation_site_regex": "",
"hic.fastq": [
[
{
"read_1": "tests/data/merged_read1.fastq.gz",
"read_2": "tests/data/merged_read2.fastq.gz"
},
],
], #From fastq
"hic.input_hic": "", #From .hic
"hic.normalization_methods": "", #VC, VC_SQRT, KR, SCALE, GW_KR, GW_SCALE, GW_VC, INTER_KR and INTER_SCALE
"hic.no_pairs": false,
"hic.no_call_loops": false, #requires GPUs
"hic.no_call_tads": false,
"hic.no_delta": false,
"hic.no_slice": false, #requires GPUs
"hic.no_eigenvectors": false,
"hic.align_num_cpus": 32,
"hic.align_ram_gb_in_situ": 64,
"hic.align_ram_gb_intact": 88,
"hic.align_disk_size_gb_in_situ": 1000,
"hic.align_disk_size_gb_intact": 1500,
"hic.chimeric_sam_nonspecific_disk_size_gb": 7000,
"hic.chimeric_sam_specific_disk_size_gb": 1500,
"hic.dedup_ram_gb_in_situ": 32,
"hic.dedup_ram_gb_intact": 48,
"hic.dedup_disk_size_gb_in_situ": 5000,
"hic.dedup_disk_size_gb_intact": 7500,
"hic.create_hic_num_cpus": ,
"hic.create_hic_ram_gb": ,
"hic.create_hic_juicer_tools_heap_size_gb": ,
"hic.create_hic_disk_size_gb": ,
"hic.add_norm_num_cpus": ,
"hic.add_norm_ram_gb": ,
"hic.add_norm_disk_size_gb": ,
"hic.create_accessibility_track_ram_gb": ,
"hic.create_accessibility_track_disk_size_gb":
}
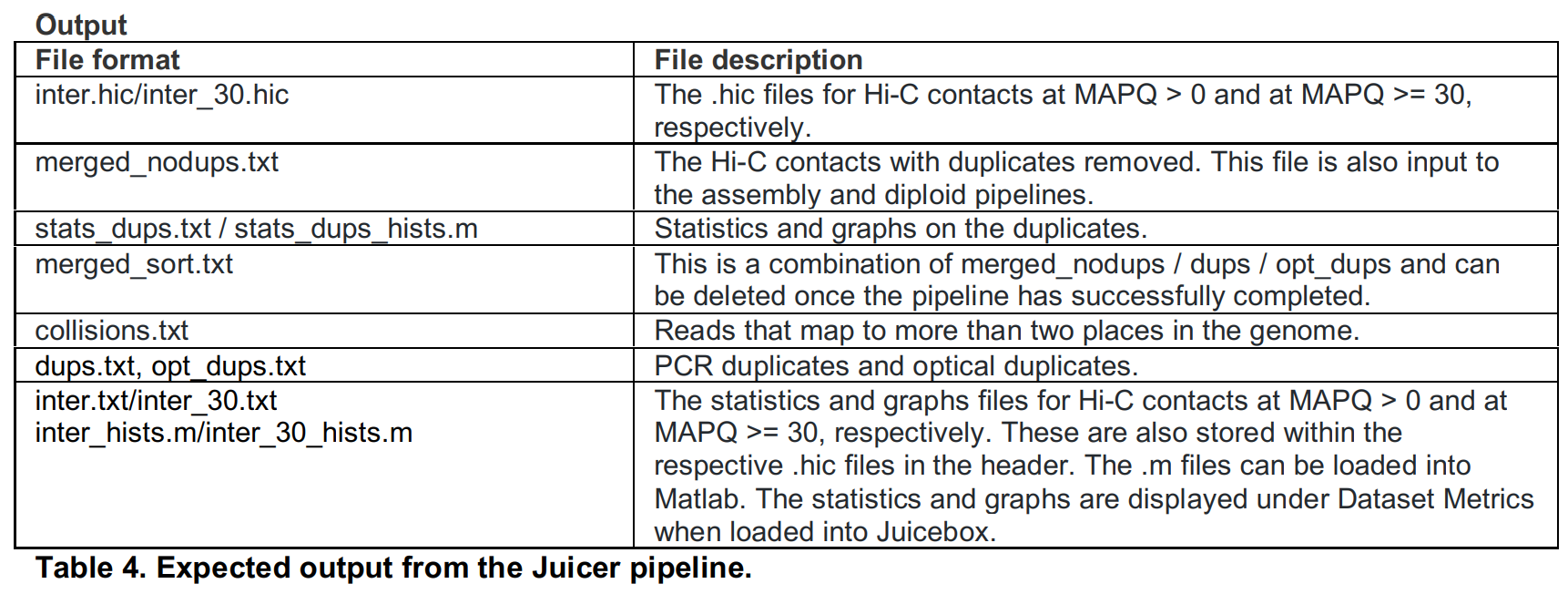
- 输出文件
首次运行会在工作目录下生成hic目录,此后在同一目录下的计算,会在hic目录下生成随机序列的目录,内存放运行结果文件,相关文件如下:
| task | outfiles | description |
|---|---|---|
| get_ligation_site_regex | ligation_site_regex.txt | 酶切位点 |
| normalize_assembly_name | normalized_assembly_name.txt, is_supported.txt | 参考基因组标识,如GRCm38 |
| align | aligned.bam, ligation_count.txt | 数据比对结果文件 |
| chimeric_sam_specific | chimeric_sam_specific.bam, result_norm.txt.res.txt | 比对文件中的chimeric reads,保留的unambiguous部分? |
| merge | merged.bam | 合并比对结果文件,包括R1和R2的正常比对以及部分的unambiguous chimeric read |
| dedup | merged_dedup.bam | 去除PCR重复后的比对文件 |
| bam_to_pre | merged_nodups_~{quality}.txt.gz, merged_nodups_~{quality}_index.txt.gz | 生成.hic文件的中间文件,详见Juicer-Pre,考虑了fragment跨越酶切位置信息,保留跨越至少一个位置,即R1和R2分布在两个或以上酶切片段 |
| pre_to_pairs | pairix.bsorted.pairs.gz | - |
| calculate_stats | stats_~{quality}.txt, stats_~{quality}.json, stats_~{quality}_hists.m | 关于比对结果的数据统计信息,默认保留MAPQ>0和MAPQ>30两种结果 |
| create_hic | inter_~{quality}_unnormalized.hic | 生成原始.hic文件,及未经过标准化 |
| add_norm | inter_~{quality}.hic | 标准化的.hic文件 |
| arrowhead | _~{quality}.bedpe.gz | Arrowhead Algorithm for Domain Annotation,检测TADs |
| hiccups | merged_loops_~{quality}.bedpe.gz | Hi-C Computational Unbiased Peak Search for Peak Calling,检测chromatin loops |
| hiccups_2 | merged_loops_~{quality}.bedpe.gz | - |
| localizer | localized_loops_~{quality}.bedpe.gz | - |
| delta | predicted_loops_merged.bedpe.gz, predicted_domains_merged.bedpe.gz, predicted_stripes_merged.bedpe.gz, predicted_loop_domains_merged.bedpe.gz | 检测TAD、loops,参考Delta |
| create_eigenvector | eigenvector_~{resolution}.wig, eigenvector_~{resolution}.bw | Compartment A is highly enriched for open chromatin; compartment B is enriched for closed chromatin,分析区别compartment A和compartment B |
| slice | slice_subcompartment_clusters_~{resolution}.bed.gz | - |
| create_accessibility_track | inter_30.bw | - |
- 问题集合
-
docker: permission denied while trying to connect to the Docker daemon socket at unix:///var/run/docker.sock
解决方法:将用户加入docker组,
sudo gpasswd -a username docker;
newgrp docker -
Unable to find image ‘encodedcc/hic-pipeline@xxx’ locally
可能是因为网络原因,无法直接从docker hub pull相关的image,
解决方法:
1)创建本地目录encodedcc,将docker目录中的镜像文件Dockerfile进行复制,
cp -r docker/ encodedcc/
这样的话好像首次运行会根据Dockerfile创建相应的容器,会耗费一定时间。 2)也可自行创建容器,
cd docker/hic-pipeline
docker build -t encodedcc/hic-pipeline:1.15.1 . -
注意,hic.reference_index提供文件为tar.gz格式,包括参考基因组bwa的index文件及序列文件
-
Hiccups报错,java.lang.UnsatisfiedLinkError: Error while loading native library “JCudaDriver-0.8.0-linux-x86_64”。
从JuicerTools下载libJCudaDriver-linux-x86_64.so库文件,将其命名为libJCudaDriver-0.8.0-linux-x86_64.so,并放在/opt/sos目录下cd /opt/soswget https://github.com/aidenlab/JuicerTools/raw/master/lib/jcuda/libJCudaDriver-linux-x86_64.so
ln -s libJCudaDriver-linux-x86_64.so libJCudaDriver-0.8.0-linux-x86_64.so
export LD_LIBRARY_PATH=$LD_LIBRARY_PATH:/opt/sos
java -jar /opt/scripts/common/juicer_tools.jar hiccups -c 19 inter_30.hic loops, -
Error response from daemon: could not select device driver “” with capabilities 这是由于没有配置好docker运行环境导致的,在container中运行GPU的环境配置方式详见docker安装步骤。
-
配置文件中fastq文件均为统一实验条件下的重复,包括生物学重复和技术从重复;对于不同实验条件下的数据,应分开运行。
HiC数据说明
参考Data Production and Processing Standard of the Hi-C Mapping Center
Quality Control
- Hi-C Library Statistics
- Aggregate Peak Analysis (APA) for Peak Calling Quality Control
- Experimental validation
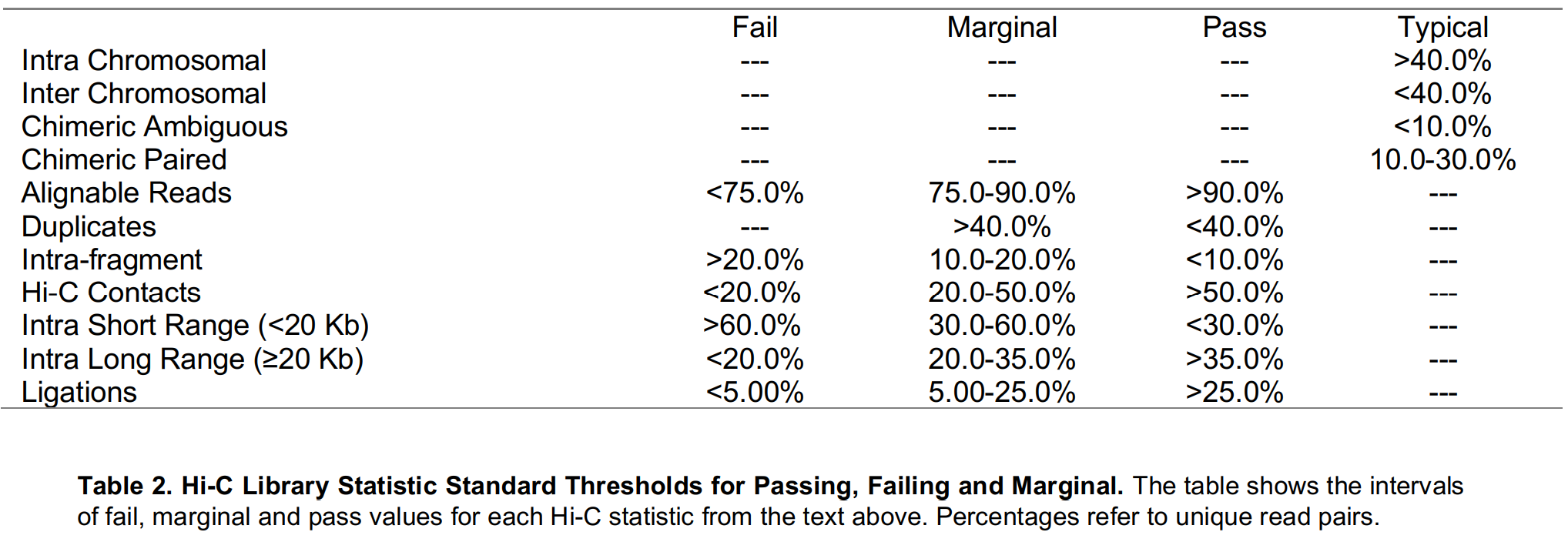
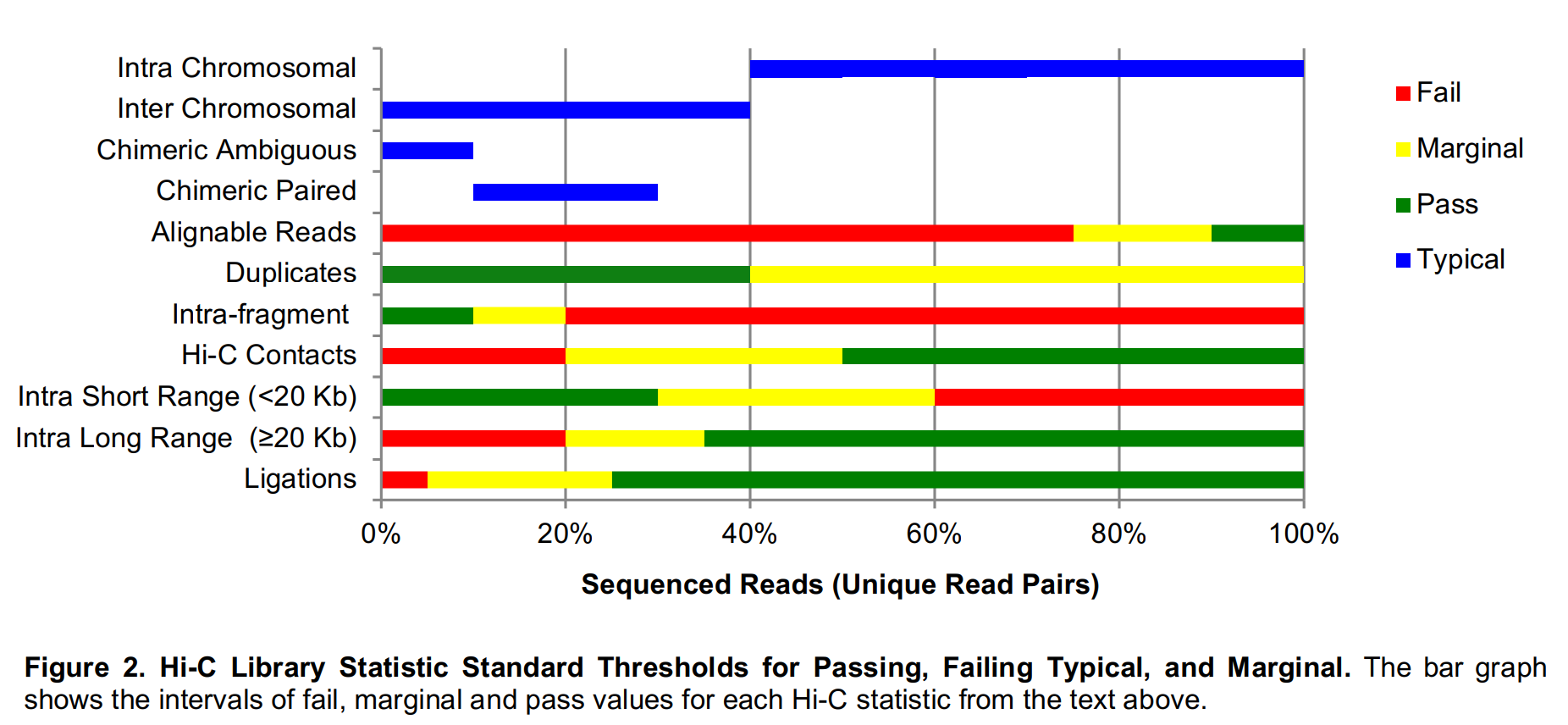
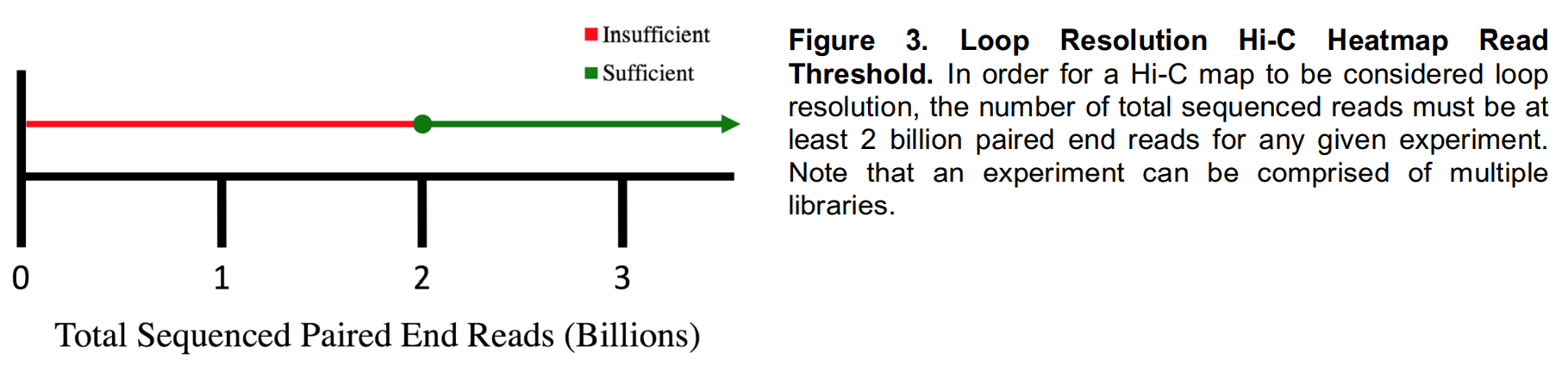
Hi-C Data Processing
Juicer pipeline consists of two main parts:
- the transformation of raw reads into a highly compressed binary file that provides fast random access to contact matrices stored at multiple resolutions with multiple normalization schemes;
- the analysis portion, which operates on the binary file and automatically annotates contact domains, loops, anchors, and compartments.
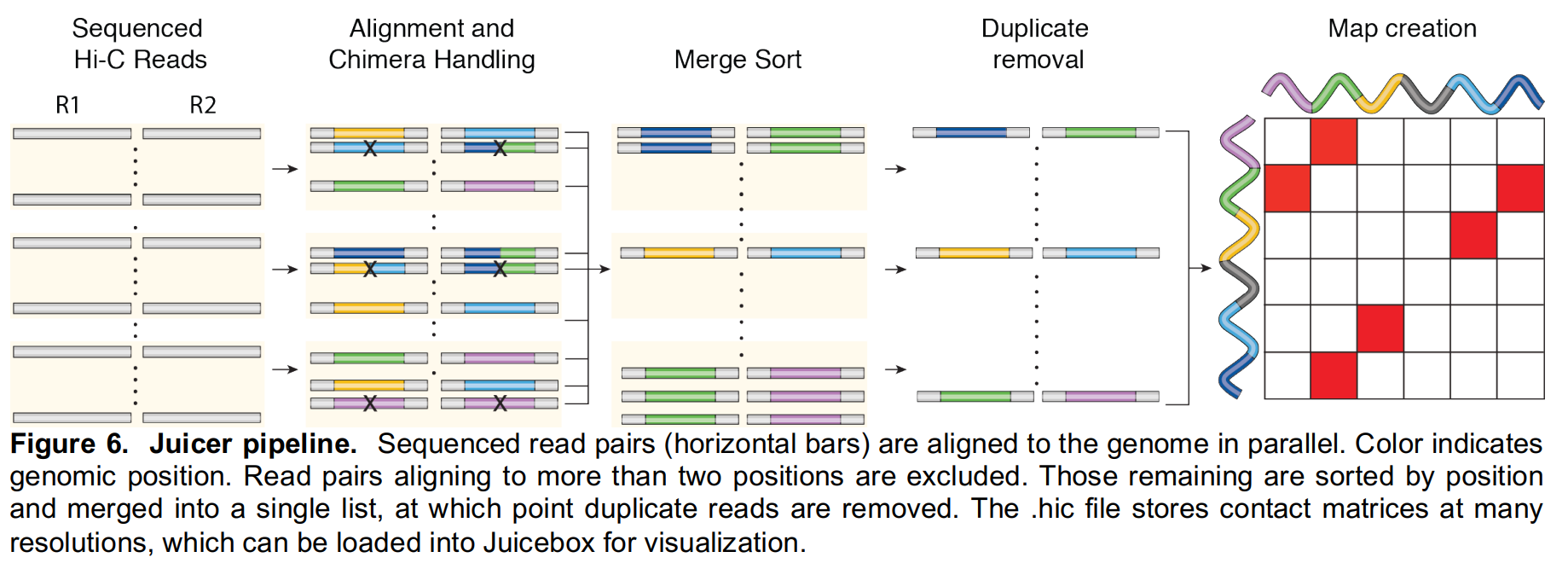
- Fastqs to contact matrices
-
Sequence alignment
1.1 Filtering of abnormal alignments
normal: each read in a read pair will align to a single site in the genome.
chimeric: at least one of the two reads comprises multiple subsequences, each of which align to different parts of the genome. For instance, the first 50 base pairs might map perfectly to one position, whereas the next 50 map perfectly to a second position several megabases away. Chimeric read pairs are classified as “unambiguous” or “ambiguous”, ambiguous chimeric reads are not included in Hi-C maps.
unalignable: they have at least one end that cannot be successfully aligned.
1.2 Filtering of duplicates
1.3 Filtering of low-quality alignments
1.4 Construction of contact matrices
默认bin大小包括2.5 Mb, 1 Mb, 500 kb, 250 kb, 100 kb, 50 kb, 25 kb, 10 kb, and 5 kb,可通过参数设置 -
Contact Matrix Normalization
2.1 Vanilla coverage normalization (VC normalization)
2.2 A role for matrix balancing in Hi-C
2.3 New methods for matrix balancing -
Diploid and Higher Order Maps
3.1 Construction of Diploid Hi-C maps
3.2 Higher-order Contact Analysis
- Feature Annotation and Other Downstream Analysis Methods
- HiCCUPS: Hi-C Computational Unbiased Peak Search for Peak Calling
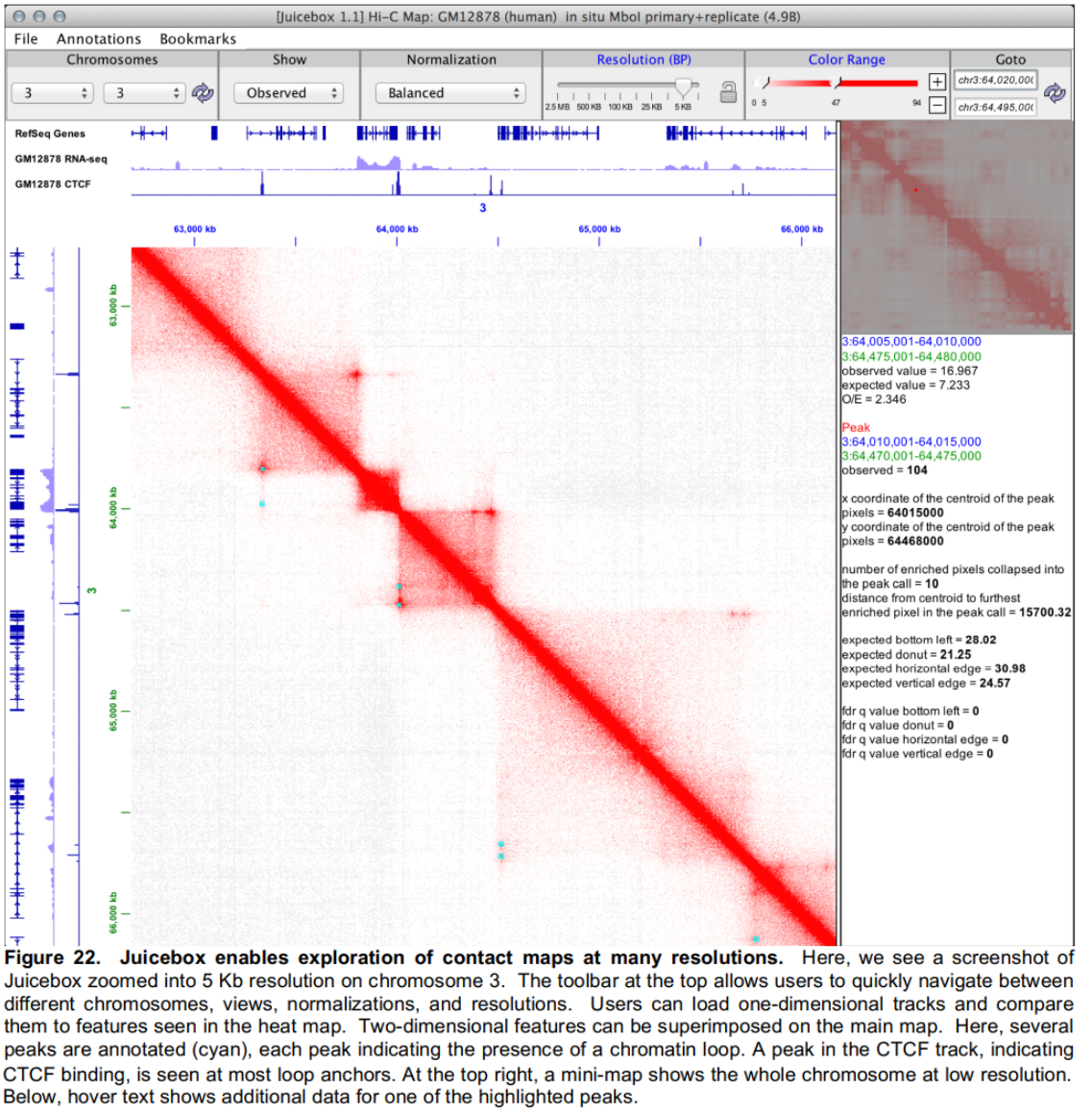
Much of the work on genome architecture so far has centered on the study of chromatin looping. Chromatin loops manifest as local peaks in a proximity ligation dataset, which occur between two points whenever they interact with each other significantly more than with random points in their neighborhood.
- Visualization and Data Integration
- Juicebox for Data Visualization and Integration with ENCODE tracks
参考基因组
##下载序列文件和注释文件
#对于人类和小鼠,从GENCODE (https://www.gencodegenes.org/) 下载参考基因组序列文件和注释文件
#Genome assembly GRCh38 /2023-02-08
wget -c https://ftp.ebi.ac.uk/pub/databases/gencode/Gencode_human/release_43/GRCh38.primary_assembly.genome.fa.gz #-c,断点续传
wget https://ftp.ebi.ac.uk/pub/databases/gencode/Gencode_human/release_43/gencode.v43.annotation.gff3.gz
wget https://ftp.ebi.ac.uk/pub/databases/gencode/Gencode_human/release_43/gencode.v43.annotation.gtf.gz
#Genome assembly GRCm38 /2020-04-29
wget -c https://ftp.ebi.ac.uk/pub/databases/gencode/Gencode_mouse/release_M25/GRCm38.primary_assembly.genome.fa.gz
wget https://ftp.ebi.ac.uk/pub/databases/gencode/Gencode_mouse/release_M25/gencode.vM25.annotation.gff3.gz
wget https://ftp.ebi.ac.uk/pub/databases/gencode/Gencode_mouse/release_M25/gencode.vM25.annotation.gtf.gz
##构建index文件
#GRCh38
bwa index -p GRCh38.primary_assembly.genome.fa.gz -a bwtsw /home/lxh/HiC_test/refgenomes/sequences/GRCh38/GRCh38.primary_assembly.genome.fa
#GRCm38
bwa index -p GRCm38.primary_assembly.genome.fa.gz -a bwtsw /home/lxh/HiC_test/refgenomes/sequences/GRCm38/GRCm38.primary_assembly.genome.fa
#染色体大小
samtools faidx GRCh38.primary_assembly.genome.fa
cut -f1,2 GRCh38.primary_assembly.genome.fa.fai > GRCh38.chrom.sizes
samtools faidx GRCm38.primary_assembly.genome.fa
cut -f1,2 GRCm38.primary_assembly.genome.fa.fai > GRCm38.chrom.sizes
##酶切片段文件
#HindIII
----------------
5' A^AGCTT 3'
3' TTCGA^A 5'
----------------
#BglII
----------------
5' A^GATCT 3'
3' TCTAG^A 5'
----------------
#DpnII
----------------
5' ^GATC 3'
3' CTAG^ 5'
----------------
#MboI
----------------
5' ^GATC 3'
3' CTAG^ 5'
----------------
##juicer
# 'HindIII' : 'AAGCTT',
# 'DpnII' : 'GATC',
# 'MboI' : 'GATC',
# 'Sau3AI' : 'GATC',
# 'Arima' : [ 'GATC', 'GANTC' ]
mkdir enzymesizes && cd enzymesizes
nano restriction_site_locations.json
caper run ../make_restriction_site_locations.wdl -i restriction_site_locations.json --docker
#restriction_site_locations.json
{
"make_restriction_site_locations.reference_fasta": "/home/lxh/HiC_test/refgenomes/sequences/GRCm38/GRCm38.primary_assembly.genome.fa",
"make_restriction_site_locations.restriction_enzyme": "DpnII",
"make_restriction_site_locations.assembly_name": "GRCm38"
}
可从ENCODE下载相关文件:
| reference file | assembly | ENCODE portal link |
|---|---|---|
| bwa index | GRCh38 | link |
| genome fasta | GRCh38 | link |
| chromosome sizes | GRCh38 | link |
| bwa index | mm10 | link |
| chromosome sizes | mm10 | link |
注意:NCBI-RefSeq、NCBI-GenBank、GENCODE、UCSC、Ensembl、ENCODE各个数据库的参考基因组对染色体的标识并不完全一样,以小鼠mm10 Chromosome1为例,GenBank: CM000994.2, RefSeq: NC_000067.6, GENCODE: chr1, UCSC: chr1, Ensembl: 1, ENCODE: chr1,此外对于随机序列,UCSC、ENCODE会标明染色体号,如chr1_GL456210_random,但GENCODE、Ensembl不会标明,如GL456210.1。下载时序列文件和注释文件应来自同一数据库。
一次hic数据分析过程
- 数据信息
物种:小鼠
| 样本名 | 实验条件 | 数据 |
|---|---|---|
| EKO4d-1 | KO | EKO4d-1_R1.fq.gz, EKO4d-1_R2.fq.gz |
| EKO4d-2 | KO | EKO4d-2_R1.fq.gz, EKO4d-2_R2.fq.gz |
| WT4d-1 | WT | WT4d-1_R1.fq.gz, WT4d-1_R2.fq.gz |
| WT4d-2 | WT | WT4d-2_R1.fq.gz, WT4d-2_R2.fq.gz |
- 质控
conda activate hicenv #激活conda环境
conda install -c bioconda multiqc
cd PATH_TO_RAW_DATA
mkdir fastqc
fastqc -o fastqc/ -t 24 ./*.fq.gz
multiqc .
- 运行hic分析流程
#准备参考文件<JuicerDIR>
#Juicer 支持UCSC或者ENCODE的参考序列文件
mkdir -p references && cd references/mm10
wget -c https://www.encodeproject.org/files/ENCFF018NEO/@@download/ENCFF018NEO.tar.gz
wget https://www.encodeproject.org/files/mm10_no_alt.chrom.sizes/@@download/mm10_no_alt.chrom.sizes.tsv
tar -xzvf ENCFF018NEO.tar.gz -C .
nano restriction_site_locations.json
caper run ../../make_restriction_site_locations.wdl -i restriction_site_locations.json --docker
#run caper<workDir>
mkdir hic_run && cd hic_run
nano input.json #json配置文件
caper run /home/lxh/HiC_test/hic-pipeline/hic.wdl -i input.json --docker
#
nohup caper run /home/lxh/HiC_test/hic-pipeline/hic.wdl -i input_EKO4d.json --docker >nohup_EKO4d.out 2>&1 &
nohup caper run /home/lxh/HiC_test/hic-pipeline/hic.wdl -i input_WT4d.json --docker >nohup_WT4d.out 2>&1 &
#hiccups分析|docker容器运行
cp /home/lxh/Lab/ZheLiu/Hi-C/hic_run/hic/fee97d45-1a06-4a50-94ee-a1db98fbf854/call-add_norm/shard-1/execution/inter_30.hic .
docker run -it -v /home/lxh/Lab/ZheLiu/Hi-C/hic_run/hic/fee97d45-1a06-4a50-94ee-a1db98fbf854/hiccups/:/shared_folder --gpus all encodedcc/hic-pipeline:1.15.1_hiccups /bin/bash
nvidia-smi #GPU信息
#java -jar /opt/scripts/common/juicer_tools.jar hiccups -m 1024 inter_30.hic loops
java -jar /opt/scripts/common/juicer_tools.jar hiccups \
-m 1024 \
-r 5000,10000,25000,50000,100000 \
-f .1,.1,.1,.1,.1 \
-p 4,2,1,1,1 \
-i 7,5,3,3,3 \
-t 0.02,1.5,1.75,2,2 \
-d 20000,20000,50000,50000,50000 \
inter_30.hic loops_2
JSON文件如下:
restriction_site_locations.json:
{
"make_restriction_site_locations.reference_fasta": "/home/lxh/HiC_test/hic-pipeline/references/mm10_no_alt_analysis_set_ENCODE.fa",
"make_restriction_site_locations.restriction_enzyme": "DpnII",
"make_restriction_site_locations.assembly_name": "mm10"
}
input.json:
{
"hic.assembly_name": "mm10",
"hic.chrsz": "/home/lxh/HiC_test/hic-pipeline/references/mm10/mm10_no_alt.chrom.sizes.tsv",
"hic.fastq": [
[
{
"read_1": "/home/lxh/Lab/ZheLiu/Hi-C/EKO4d-1_R1.fq.gz",
"read_2": "/home/lxh/Lab/ZheLiu/Hi-C/EKO4d-1_R2.fq.gz"
},
{
"read_1": "/home/lxh/Lab/ZheLiu/Hi-C/EKO4d-2_R1.fq.gz",
"read_2": "/home/lxh/Lab/ZheLiu/Hi-C/EKO4d-2_R2.fq.gz"
}
],
[
{
"read_1": "/home/lxh/Lab/ZheLiu/Hi-C/WT4d-1_R1.fq.gz",
"read_2": "/home/lxh/Lab/ZheLiu/Hi-C/WT4d-1_R2.fq.gz"
},
{
"read_1": "/home/lxh/Lab/ZheLiu/Hi-C/WT4d-2_R1.fq.gz",
"read_2": "/home/lxh/Lab/ZheLiu/Hi-C/WT4d-2_R2.fq.gz"
}
]
],
"hic.reference_index": "/home/lxh/HiC_test/hic-pipeline/references/mm10/ENCFF018NEO.tar.gz",
"hic.restriction_enzymes": [
"DpnII"
],
"hic.restriction_sites": "/home/lxh/HiC_test/hic-pipeline/references/mm10/mm10_DpnII.txt.gz"
}
#input_EKO4d.json
{
"hic.assembly_name": "mm10",
"hic.chrsz": "/home/lxh/HiC_test/hic-pipeline/references/mm10/mm10_no_alt.chrom.sizes.tsv",
"hic.fastq": [
[
{
"read_1": "/home/lxh/Lab/ZheLiu/Hi-C/EKO4d-1_R1.fq.gz",
"read_2": "/home/lxh/Lab/ZheLiu/Hi-C/EKO4d-1_R2.fq.gz"
},
{
"read_1": "/home/lxh/Lab/ZheLiu/Hi-C/EKO4d-2_R1.fq.gz",
"read_2": "/home/lxh/Lab/ZheLiu/Hi-C/EKO4d-2_R2.fq.gz"
}
]
],
"hic.reference_index": "/home/lxh/HiC_test/hic-pipeline/references/mm10/ENCFF018NEO.tar.gz",
"hic.restriction_enzymes": [
"DpnII"
],
"hic.restriction_sites": "/home/lxh/HiC_test/hic-pipeline/references/mm10/mm10_DpnII.txt.gz"
}
#input_WT4d.json
{
"hic.assembly_name": "mm10",
"hic.chrsz": "/home/lxh/HiC_test/hic-pipeline/references/mm10/mm10_no_alt.chrom.sizes.tsv",
"hic.fastq": [
[
{
"read_1": "/home/lxh/Lab/ZheLiu/Hi-C/WT4d-1_R1.fq.gz",
"read_2": "/home/lxh/Lab/ZheLiu/Hi-C/WT4d-1_R2.fq.gz"
},
{
"read_1": "/home/lxh/Lab/ZheLiu/Hi-C/WT4d-2_R1.fq.gz",
"read_2": "/home/lxh/Lab/ZheLiu/Hi-C/WT4d-2_R2.fq.gz"
}
]
],
"hic.reference_index": "/home/lxh/HiC_test/hic-pipeline/references/mm10/ENCFF018NEO.tar.gz",
"hic.restriction_enzymes": [
"DpnII"
],
"hic.restriction_sites": "/home/lxh/HiC_test/hic-pipeline/references/mm10/mm10_DpnII.txt.gz"
}
- 下游分析和可视化
HiCExplorer,Set of programs to process, analyze and visualize Hi-C and cHi-C data.
